

Due to network problems, notification mails of the results may fail to be delivered to you. Once your job has been successfully executed, the results become available from the following URL: http://prokatlas.bs.s.u-tokyo.ac.jp/habitat_pref_scores_YOURJOBID.txt (replace YOURJOBID with your job id, which starts with 2024) –– if you do not receive the result after a reasonable amount of time (~15 min for up to ~100 sequences), please try directly accessing the URL above (Jan 11th, 2024).
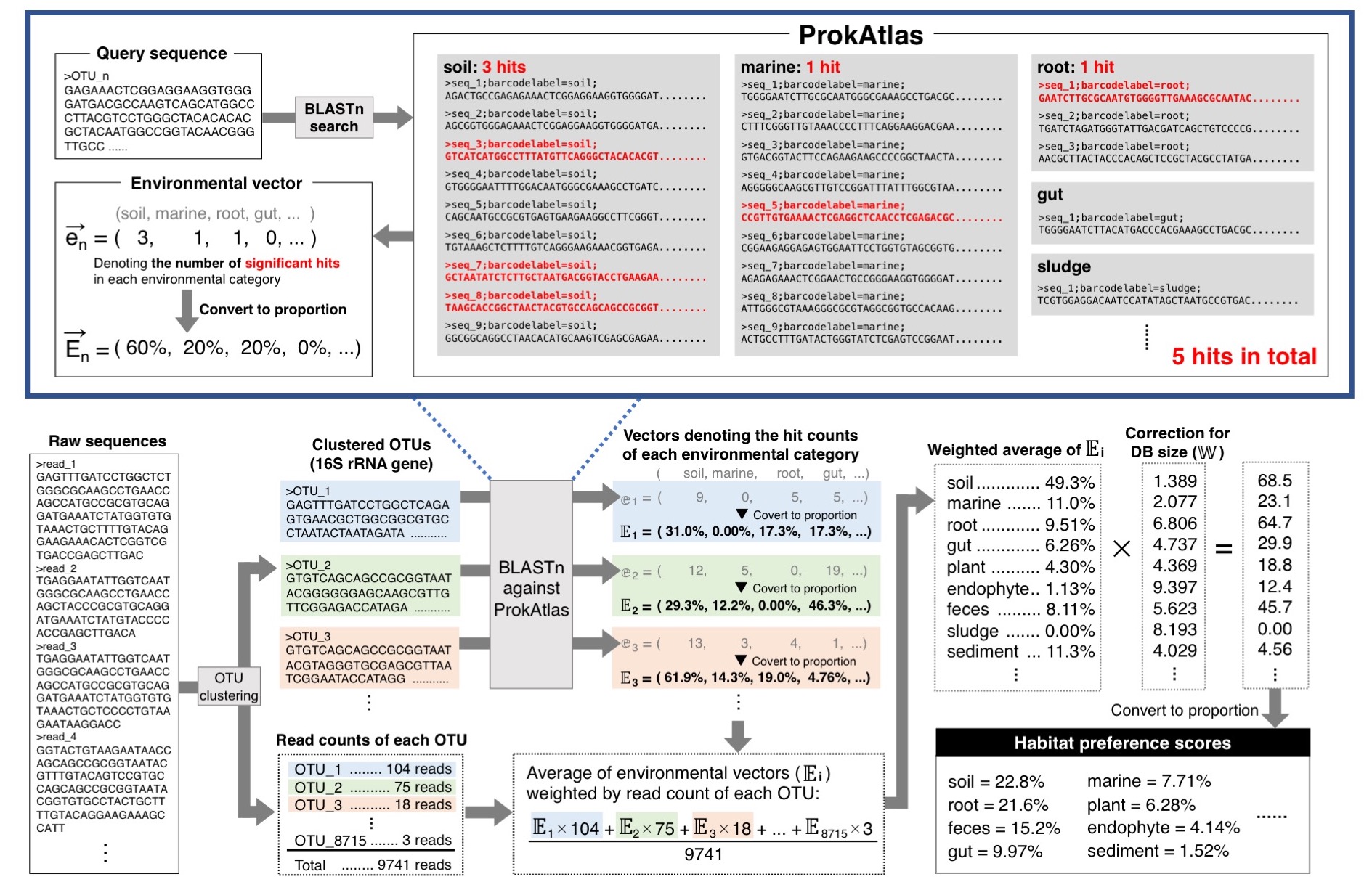
ProkAtlas is a bioinformatic database and analysis pipeline that infer the habitat environment of prokaryotes from their 16S rRNA gene sequences. This webpage, entitled ProkAtlas Online Toolkit, provides a ready-to-use web interface for analyzing prokaryotic habitat preferences.
For more details, please see the instructions available here. You can also obtain the database and source code for stand-alone analysis using ProkAtlas.
To predict the habitat preferences of individual prokaryotic species, just submit their whole or partial 16S ribosomal RNA/DNA sequence(s) in FASTA format. To calculate the habit preferences of prokaryotic communities, submit a simple tab-delimited OTU table (an example available below) along with 16S rRNA/rDNA sequences (i.e. OTU representative sequences).
While we provide a user-friendly interface below, raw ProkAtlas database and pipeline for calculating habitat preference scores, which requires only blast+, python, and pandas, are available here. Please also see our paper reporting the development and use cases of ProkAtlas.
ProkAtlas contains 361,474 sequences, each labeled by one environmental category. Any query sequence of 16S rRNA gene is subjected to BLASTn search against ProkAtlas database, and "significant" hits satisfying the alignment criteria.
In many cases, BLASTn finds multiple hits across several environmental categories, and the composition of environmental categories within those hits are calculated. This composition, in principle, denotes the habitat preference of microbes corresponding to query sequence.
By compiling this composition for each community member, overall habitat preference scores representing a microbial community can be obtained.
Although we are trying to minimize the possible errors and biases in ProkAtlas, the results are always affected by "missing data errors". Because ProkAtlas does not cover all the microbes and samples on Earth, rare habitats may often dismissed by ProkAtlas.
More details are explained in our paper.
If you used ProkAtlas in your study, please cite the following paper: